5 Setup of Solo Workflow
5.1 Outline for this section
We have discussed why Git and GitHub are important, now we will set up a repository and work through an example. During this first section, you will be working in a repository alone. We will:
- Make a new repository on GitHub
- Clone the repository to your local machine
- Write code in your repository locally
- Push the code to your repository
- Merge your branch into main
5.2 Working in Terminal (Mac) and Bash (Windows)
- We will use Terminal and Bash applications to interact with Git on our laptops.
- Below is a Mac Terminal window. It looks very similar to a Windows Bash window.
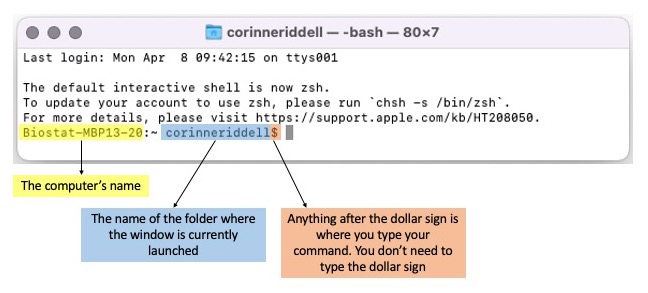
- In this session, we will supply you with Git code for you to manually input into your Terminal/Bash windows. Note that all of this code will be written after the dollar sign in the figure above.
5.3 Working in Terminal (Mac) and Bash (Windows)
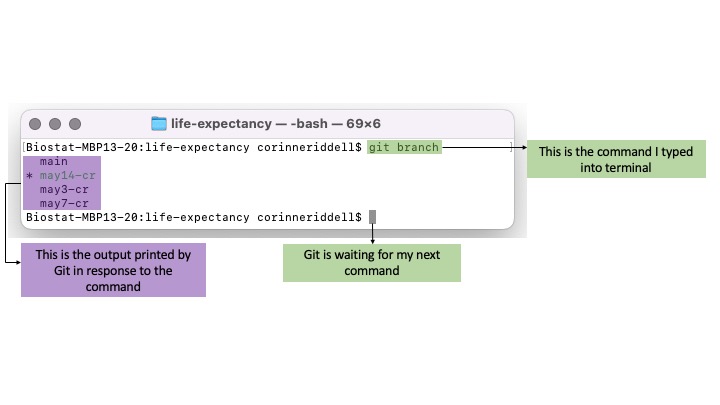
- Here is an example of the command
git branch, followed by the output printed to screen:
5.4 Folder names and file names tips
- Recall when you name a variable in SAS or R, the variable name cannot contain spaces or unusual characters.
- It is best practice to not use spaces or unusual characters in folder or file names, even though spaces are permissible and commonly used by Windows and Mac Users.
5.6 What is the problem with spaces, anyway?
- While spaces are human-readable they aren’t machine-friendly.
- When you refer to a folder or file using Git in Terminal or Bash, a name without spaces is much easier to type (otherwise you have to insert a backslash before the space)
- Spaces also break the auto-complete function that Git users love. This is a very frustrating experience.
5.7 Good file names are…
- machine readable
- human readable
- play well with default ordering
See the file “how-to-name-files.pdf” for a more deeper dive into file naming.
5.7.1 Good file names examples
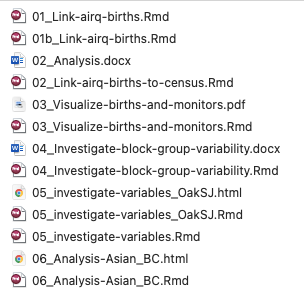
- Filenames start with a number (padded by 0) to order the files according to the order performed in the analysis
- This is followed by a short (human and machine readable) descriptor of what the file does
- Uses underscore “_” to delimit field, and dashes “-” to separate words within field
5.7.2 Bad file name example (and associated Git pain)
- Can see that there is an R markdown (Rmd) file named “Data Visualization Evaluation Report” that has been modified.
- The pain arises when I go to
git addthe file. Before each space, I need to include a backslash (which looks ugly). Even worse, the space breaks the auto-complete that happens when I press “tab” to auto-complete the file name. Auto-complete will become your friend when you use Git, and not being able to use it is very sad/infuriating when you have grown to love it.

5.8 Set up a project that you want to track with git and GitHub
Let’s suppose that you have a project that you want to start tracking using Git and Github. For this project, you are already working on with some code, data, and visualizations that have already been saved. We have made this project for you. To download it, run the following two commands in the RStudio console:
install.packages("usethis")
usethis::use_course("corinne-riddell/existing-project")- R will asked you if you want this folder copied onto the Desktop. Select Yes.
- R will display messages showing you that the folder has been downloaded and unzipped. Tell R whether to delete the file.
- RStudio will then open. Click the code folder in the file viewer. Then, click the filename “01_Analyze-life-expectancy.R” to open this file in RStudio.
- Run all the code in the .R file you just downloaded. Note that it created a figure and saved it into the images sub-folder.
Now you are set up with some existing code and things that you might want to start tracking on GitHub. The next thing to do is make a folder on GitHub that will store this project.
5.9 Make a new repository on GitHub
- Go to github.com and log in. Click the green “New” button to make a GitHub repository. Type “life-expectancy” in the repository name.
- Write whatever you want in the description. For example, type “An analysis of life expectancy in the US”
- Select either to make this a public or private repository.
- Check the box next to
Add a README file. This tells Git to create a file that can describe your project. For now, you can write a sentence about this being a practice repository for this workshop.
- Choose
.gitignoretemplate:R. This tells Git to use defaults that work well for R users.
- Choose
MIT License. This relates to the licensing for your code, which is not relevant for this workshop.
- Click “Create repository”. Github will then bring you to the repository’s main page.
5.10 Clone the repository to your local machine
- From the main page of your repository and click on the green
Codebutton.
- You’ll see a URL that starts with https://. Push the icon with two overlapping squares to copy the URL to your clipboard. [Lauren, you had written this differently and had them use the SSH one… is there a reason to use one over the other?]
- Open terminal (Mac) or bash (Windows) program. Navigate to where you want to place
this repository using the
cd {folder_name}command. Then writegit clone {paste the url you copied here}and then press the return/enter button. The following will display in Terminal/Bash if this was successful:
Biostat-MBP13-20:repos corinneriddell$ git clone https://github.com/corinne-riddell/life-expectancy.git
Cloning into 'life-expectancy'...
remote: Enumerating objects: 5, done.
remote: Counting objects: 100% (5/5), done.
remote: Compressing objects: 100% (5/5), done.
remote: Total 5 (delta 0), reused 0 (delta 0), pack-reused 0
Unpacking objects: 100% (5/5), done.You now are ready to begin tracking changes to this folder using Git and GitHub.
5.11 Get oriented to the new directory
To get yourself oriented, do the following in the Terminal/Bash window:
- Navigate into your repository by typing
cd life-expectancy/. - Type
git status. The results shows you that no changes have been made yet:
Biostat-MBP13-20:life-expectancy corinneriddell$git status
On branch main
Your branch is up to date with 'origin/main'.
nothing to commit, working tree clean- Type
git branch. This shows you that you are currently on the main branch.
Biostat-MBP13-20:life-expectancy corinneriddell$git branch
* main5.12 Make your first branch
Set yourself up in a new branch off of main. In terminal/bash:
- Type
git checkout -b may3-XY, replacing XY with your initials. (If today is not May 3, replace “may3” with today’s date.)
Biostat-MBP13-20:life-expectancy corinneriddell$ git checkout -b may3-cr
Switched to a new branch 'may3-cr'- Type
git branch, to confirm to yourself that you have indeed switched to the new branch.
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
main
* may3-cr5.13 Make some changes to your code
Okay, you are now set up to track changes. Let’s do the following:
- Copy the code/ data/ and images/ sub-folders from your “existing-project” folder into the “life-expectancy” folder.
5.14 Commit the changes that you made and push them to GitHub
- Go back over to terminal or bash. Type
git status. The output will tell you what has been changed. It tells us that there are untracked files:
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may3-cr
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
code/
data/
images/
life-expectancy.RprojWe want to track the code/, data/ and images/ subfolders we just copied over, as well as the life-expectancy.Rproj file.
Use git add to add the newly-added files to be tracked. Then use git status
to confirm you have added everything you want to track:
Biostat-MBP13-20:life-expectancy corinneriddell$ git add code/
Biostat-MBP13-20:life-expectancy corinneriddell$ git add data/
Biostat-MBP13-20:life-expectancy corinneriddell$ git add images/
Biostat-MBP13-20:life-expectancy corinneriddell$ git add life-expectancy.Rproj
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may3-cr
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
new file: code/01_Analyze-life-expectancy.R
new file: data/Life-expectancy-by-state-long.csv
new file: images/ca-black-women-LE.png
new file: images/placeholder.md
new file: life-expectancy.Rproj
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_StoreNote: Computers create files that we don’t want to track. For example, Macs create .DS_Store files. Another example is that Windows creates temporary files when a Word doc or Excel spreadsheet is open. You will see these weird files listed under the Untracked files list. You don’t need to worry about them because we don’t want to track changes to any of those files.
- Commit these changes locally:
git commit -m 'your commit message', replace ‘your commit message’ with a short message about what you’ve done (keep the quotes around the message). For example, your message could be something likegit commit -m "added first set of files".
Biostat-MBP13-20:life-expectancy corinneriddell$ git commit -m "added first set of files"
[may3-cr 58fcc58] added first set of files
5 files changed, 7253 insertions(+)
create mode 100644 code/01_Analyze-life-expectancy.R
create mode 100644 data/Life-expectancy-by-state-long.csv
create mode 100644 images/ca-black-women-LE.png
create mode 100644 images/placeholder.md
create mode 100644 life-expectancy.Rproj- Push these changes to GitHub:
git push origin your-branch-name, replacing your-branch-name with the name of your branch. If you don’t remember your branch’s name, typegit branchto print it to the screen and then thegit pushcommand.
Biostat-MBP13-20:life-expectancy corinneriddell$ git push origin may3-cr
Enumerating objects: 11, done.
Counting objects: 100% (11/11), done.
Delta compression using up to 8 threads
Compressing objects: 100% (9/9), done.
Writing objects: 100% (10/10), 136.50 KiB | 10.50 MiB/s, done.
Total 10 (delta 0), reused 0 (delta 0)
remote:
remote: Create a pull request for 'may3-cr' on GitHub by visiting:
remote: https://github.com/corinne-riddell/life-expectancy/pull/new/may3-cr
remote:
To https://github.com/corinne-riddell/life-expectancy.git
* [new branch] may3-cr -> may3-cr
You have successfully pushed your changes to GitHub!
5.15 Merge the changes from your branch into main
- Navigate to GitHub.com to your repository’s URL. There should be a pale yellow
banner informing you about the changes you just pushed. Click the button
“Compare & pull request”. Notice that the title is your commit message from the
previous step. Scroll down. Look at the files that have been added.
- The code is all shown in green, indicating that every line of code is new.
- The csv data file has been added but is not rendered because it is large
- The png file is displayed.
- Click on the green “Create pull request” button. Github will check that it is able to merge your branch with main without problems. Note the message “This branch has no conflicts with the base branch”. This means you are good to go!
- Click on the green “Merge pull request” button.
- Click on the green “Confirm merge” button.
- Click the “Delete branch” button.
5.16 Summary
- You setup a folder on your laptop so that Git is used to track changes made locally on your laptop.
- You linked that folder to GitHub.com so that the same changes can be tracked externally on GitHub.
- You compared changes you made locally on your branch to the main branch on GitHub and pulled your changes into main. This means that the main branch has been updated with your changes on GitHub.
5.17 Tracking changes
Now suppose a few days have gone by and you are ready to work on your analysis project. In particular, you want to update some code that will affect some of the results and “outputs”, where outputs are results saved in any form. In this section, we outline the process to follow when you want to implement some set of tracked changes.
5.18 Get setup for a new day of work
The first thing we need to do is make sure we are in a good place with git and GitHub:
- Open up bash or terminal and navigate to the life-expectancy folder using
the
cdcommand. - Check which branch you are currently on using
git branch. All local branch names are displayed. The asterisk is next to the branch we are currently on.
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
main
* may3-cr- Check if you forgot to save anything from last time using
git status. Ideally, you have saved all your changes and there is nothing to add/track/commit. Here is my status:
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may3-cr
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
nothing added to commit but untracked files present (use "git add" to track)There is one untracked file: .DS_Store. This is okay since it is an internal file used by Mac OS that does not need to be tracked. We just want to ensure no code files or outputs we intended to track have been forgotton.
- You are likely still on your branch from the last day. In that case, navigate
back to main using
git checkout main.
Biostat-MBP13-20:life-expectancy corinneriddell$ git checkout main
Switched to branch 'main'
Your branch is up to date with 'origin/main'.If you want to double check, type git branch to confirm you are on
main.
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
* main
may3-crYou can also delete the “may3-XY” branch since you no longer need to track it locally:
Biostat-MBP13-20:github-training corinneriddell$ git branch -d may3-cr
Deleted branch may3-cr (was 761bb97).CORINNE TODO: ON HOME COMPUTER DELETE THE MAY3 BRANCH FROM THE LE REPO SO CAN GRAB THE CORRECT CODE AFTER “WAS” AND REPLACE IN THE ABOVE.
5.19 Pull down the changes from main
This is your local copy of main. It needs to pull down the changes to
made that you made on GitHub in an earlier step. To do that, type git pull origin main.
A graphic will be drawn that summarizes which files have been updated and by
how much.
Biostat-MBP13-20:life-expectancy corinneriddell$ git pull origin main
remote: Enumerating objects: 1, done.
remote: Counting objects: 100% (1/1), done.
remote: Total 1 (delta 0), reused 0 (delta 0), pack-reused 0
Unpacking objects: 100% (1/1), done.
From https://github.com/corinne-riddell/life-expectancy
* branch main -> FETCH_HEAD
884028e..5f81e34 main -> origin/main
Updating 884028e..5f81e34
Fast-forward
code/01_Analyze-life-expectancy.R | 38 +
data/Life-expectancy-by-state-long.csv | 7201 +++++++++++++++++++++++++++++++++
images/ca-black-women-LE.png | Bin 0 -> 74550 bytes
images/placeholder.md | 1 +
life-expectancy.Rproj | 13 +
5 files changed, 7253 insertions(+)
create mode 100644 code/01_Analyze-life-expectancy.R
create mode 100644 data/Life-expectancy-by-state-long.csv
create mode 100644 images/ca-black-women-LE.png
create mode 100644 images/placeholder.md
create mode 100644 life-expectancy.Rproj5.20 Start a new branch
- Like the last day, start a new branch to track today’s changes. Let’s
pretend it is now May 7. Type
git checkout -b may7-XY, where XY is replaced with your initials. Typegit branchto confirm you have changed branches.
Biostat-MBP13-20:life-expectancy corinneriddell$ git checkout -b may7-cr
Switched to a new branch 'may7-cr'
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
main
* may7-cr5.21 You are now ready to make changes!
Think about what you would like to do in advance. In particular, suppose you want to:
make a table that summarizes the mean life expectancy by race and gender for each state and,
save the above table as a CSV file into the data folder.
Re-launch RStudio by double-clicking the .Rproj file in your file viewer window. Navigate to the code file 01_Analyze-life-expectancy.R and insert the following R code (if working in R) or SAS code (if working in SAS) to make and save this table:
# R Code
# Calculate the LE for each state, separately by race and gender:
le_averages <- le_data %>%
group_by(state, race, sex) %>%
summarise(mean_LE = mean(LE))
# print the first 10 rows to the screen. By default, R rounds the numeric
# information in the display to make it more compact
le_averages
# alternatively, type View(le_averages) in the Console to open up a Viewer
# window, or click the table icon beside the le_averages objects in the
# Environment pane (upper right hand panel of RStudio).
#save this table as a CSV file in the data sub-folder
write_csv(le_averages, "./data/le_averages.csv")/*SAS Code*/
/*Calculate the LE for each state, separately by race and gender:*/
proc sort data=le_data; by state race sex; run;
proc means data=le_data; by state race sex; var le; output out=le_averages mean=mean_le ; run;
/*Print the first 10 rows to the screen*/
proc print data=le_averages (obs=10); run;
/*Or you could just open the dataset to browse it.*/
/*Export this file to a .csv file
(if you use the following code, don't forget to replace YourFilePathHere with the appropriate file path!)*/
PROC EXPORT DATA= WORK.LE_AVERAGES
OUTFILE= "YourFilePathHere\data\le_averages.csv"
DBMS=CSV REPLACE;
PUTNAMES=YES;
RUN;5.22 Track your changes using Git
Re-run your previous R code (highlight all the previous code and hit command + Return [Mac] or control + Enter [Windows]). Then run the newly-added code line by line to see what it is doing. Save the updated .R file by pushing the save icon.
Track the changes using Git. Go to the bash/terminal window. Type “git status”. Which files have been modified? Which files are new and untracked?
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may7-cr
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: code/01_Analyze-life-expectancy.R
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
data/le_averages.csv5.23 Add new and modified files
- Use
git addto add the specific files that have been modified or created. Add them one by one. Usegit statusagain to check that all the changed files are being tracked. When you are satisfied, commit these changes locally.
Biostat-MBP13-20:life-expectancy corinneriddell$ git add code/01_Analyze-life-expectancy.R
Biostat-MBP13-20:life-expectancy corinneriddell$ git add data/le_averages.csv
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may7-cr
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
modified: code/01_Analyze-life-expectancy.R
new file: data/le_averages.csv
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store5.24 Commit the changes locally
Helpful hint: Terminal/Bash plays well with autocomplete. For example, if you are typing the pathway for the .R file as “code/…” you can push the tab button as you are typing the name and it will autocomplete. This makes selecting the specific files to commit much easier.
- “git commit -m ‘your message’”. Replace ‘your message’ with a short message describing the changes. Remember to keep the quotes around the message!
Biostat-MBP13-20:life-expectancy corinneriddell$ git commit -m 'calc LE averages'
[may7-cr a7435b8] calc LE averages
2 files changed, 177 insertions(+), 1 deletion(-)
create mode 100644 data/le_averages.csv5.25 Push the changes up to GitHub
- You are now ready to push these changes up to GitHub onto GitHub’s version
of your local branch. First, remind yourself of your branch’s name using
“git branch”. Then push:
git push origin {YOUR-BRANCH-NAME}, replacing {YOUR-BRANCH-NAME} with the name of your branch.
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
main
* may7-cr
Biostat-MBP13-20:life-expectancy corinneriddell$ git push origin may7-cr
Enumerating objects: 10, done.
Counting objects: 100% (10/10), done.
Delta compression using up to 8 threads
Compressing objects: 100% (6/6), done.
Writing objects: 100% (6/6), 2.84 KiB | 2.84 MiB/s, done.
Total 6 (delta 2), reused 0 (delta 0)
remote: Resolving deltas: 100% (2/2), completed with 2 local objects.
remote:
remote: Create a pull request for 'may7-cr' on GitHub by visiting:
remote: https://github.com/corinne-riddell/life-expectancy/pull/new/may7-cr
remote:
To https://github.com/corinne-riddell/life-expectancy.git
* [new branch] may7-cr -> may7-cr5.26 Create a pull request
- Navigate to GitHub.com and go through the steps described previously to create a pull request to pull these changes into main.
5.27 Changes after receiving an updated dataset
Another week goes by. It is now May 14. You received an email that there was an error in the data file that you used to conduct the analysis. A new data file was securely transferred to you by the data holder. You need to rerun the analysis using the new dataset. The new data file is the one called “LEbsyrx.csv” in the data folder.
5.28 Get ready for the day
Set yourself up to work with git and GitHub for the day:
git branch: see which branch you are on.
Biostat-MBP13-20:life-expectancy corinneriddell$ git branch
main
* may7-cr-
git status: confirm you committed everything you wanted to commit.
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may7-cr
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
nothing added to commit but untracked files present (use "git add" to track)-
git checkout main: switch to the main branch -
git branch -d may7-cr: delete the old branch (change “cr” to your initials)
Biostat-MBP13-20:life-expectancy corinneriddell$ git checkout main
Switched to branch 'main'
Your branch is up to date with 'origin/main'.-
git pull origin main: pull GitHub’s copy of the main branch to update your local version. Examine the figure made by git about the changes.
Biostat-MBP13-20:life-expectancy corinneriddell$ git pull origin main
remote: Enumerating objects: 1, done.
remote: Counting objects: 100% (1/1), done.
remote: Total 1 (delta 0), reused 0 (delta 0), pack-reused 0
Unpacking objects: 100% (1/1), done.
From https://github.com/corinne-riddell/life-expectancy
* branch main -> FETCH_HEAD
5f81e34..e15298a main -> origin/main
Updating 5f81e34..e15298a
Fast-forward
code/01_Analyze-life-expectancy.R | 17 ++++-
data/le_averages.csv | 161 +++++++++++++++++++++++++++++++++++++++
2 files changed, 177 insertions(+), 1 deletion(-)
create mode 100644 data/le_averages.csv5.29 Checkout a new branch
-
git checkout -b may14-xy, replacing xy with your initials.
Biostat-MBP13-20:life-expectancy corinneriddell$ git checkout -b may14-cr
Switched to a new branch 'may14-cr'5.30 Update the dataset
Update the CSV file “Life-expectancy-by-state-long.csv” with the new dataset. First, decide if you want to archive this older version of the dataset for any reason. If you do, then decide where you would store the archived version and move it there. The archived version could stay on GitHub or be moved off of GitHub – this is up to you and your file organization system.
Move the LEbsyrx.csv into the data folder and rename it to have the name of the file it is replacing (“Life-expectancy-by-state-long.csv”).
5.31 Re-run the analysis
- Re-run all your code that uses this file. First open the .Rproj file to launch RStudio. Then, you can highlight all the code and hit the “Run” button.
5.32 Track the changes using Git
Use “git status”, “git add…”, and “git commit…” to track these changes locally. Use “git push” to push these changes to GitHub.
Using git status, I can see that the data files have all been modified, as has
the image file of the plot. There is a new untracked folder called data/archive/
where I moved the archived dataset.
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may14-cr
Changes not staged for commit:
(use "git add <file>..." to update what will be committed)
(use "git restore <file>..." to discard changes in working directory)
modified: data/Life-expectancy-by-state-long.csv
modified: data/le_averages.csv
modified: images/ca-black-women-LE.png
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
data/.DS_Store
data/archive/I use git add to specify all the new things I want to track:
Biostat-MBP13-20:life-expectancy corinneriddell$ git add data/Life-expectancy-by-state-long.csv
Biostat-MBP13-20:life-expectancy corinneriddell$ git add data/le_averages.csv
Biostat-MBP13-20:life-expectancy corinneriddell$ git add images/ca-black-women-LE.png
Biostat-MBP13-20:life-expectancy corinneriddell$ git add data/archive/I then use git status to confirm everything is being tracked:
Biostat-MBP13-20:life-expectancy corinneriddell$ git status
On branch may14-cr
Changes to be committed:
(use "git restore --staged <file>..." to unstage)
modified: data/Life-expectancy-by-state-long.csv
new file: data/archive/Life-expectancy-by-state-long_old.csv
modified: data/le_averages.csv
modified: images/ca-black-women-LE.png
Untracked files:
(use "git add <file>..." to include in what will be committed)
.DS_Store
data/.DS_Store5.33 Commit the changes locally
Then I commit my changes:
Biostat-MBP13-20:life-expectancy corinneriddell$ git commit -m 'data update and downstream changes'
[may14-cr c4b02db] data update and downstream changes
4 files changed, 7570 insertions(+), 369 deletions(-)
create mode 100644 data/archive/Life-expectancy-by-state-long_old.csv
rewrite images/ca-black-women-LE.png (98%)5.34 Push the changes to GitHub
Finally I push these changes to GitHub:
git push origin may14-cr
Enumerating objects: 14, done.
Counting objects: 100% (14/14), done.
Delta compression using up to 8 threads
Compressing objects: 100% (8/8), done.
Writing objects: 100% (8/8), 68.08 KiB | 9.73 MiB/s, done.
Total 8 (delta 3), reused 0 (delta 0)
remote: Resolving deltas: 100% (3/3), completed with 3 local objects.
To https://github.com/corinne-riddell/life-expectancy.git
e15298a..c4b02db may14-cr -> may14-cr
Biostat-MBP13-20:life-expectancy corinneriddell$
5.35 Submit a pull request
- Go through the process to start a pull request. On the pull request page, scroll down to see the “diffs” in the data and image. Pay close attention to the files that were changed:
- use the “2-up” “swiper” and “onion skin” tools to see the changes to the saved figure. How did the data changed? Which tool do you prefer?
- can you see which rows of data were affected by the change?
- can you tell from the data or images how the change affected the analytic findings?
- anything else you noticed?